Generate a matrix describing the amount of shared boundary length between different planning units, and the total amount of boundary length for each planning unit.
Usage
boundary_matrix(x, ...)
# S3 method for class 'Raster'
boundary_matrix(x, ...)
# S3 method for class 'SpatRaster'
boundary_matrix(x, ...)
# S3 method for class 'SpatialPolygons'
boundary_matrix(x, ...)
# S3 method for class 'SpatialLines'
boundary_matrix(x, ...)
# S3 method for class 'SpatialPoints'
boundary_matrix(x, ...)
# S3 method for class 'sf'
boundary_matrix(x, ...)
# Default S3 method
boundary_matrix(x, ...)Arguments
- x
terra::rast()orsf::sf()object representing planning units.- ...
not used.
Value
A Matrix::dsCMatrix symmetric sparse matrix object.
Each row and column represents a planning unit.
Cell values indicate the shared boundary length between different pairs
of planning units. Values along the matrix diagonal indicate the
total perimeter associated with each planning unit.
Details
This function assumes the data are in a coordinate
system where Euclidean distances accurately describe the proximity
between two points on the earth. Thus spatial data in a
longitude/latitude coordinate system (i.e.,
WGS84)
should be reprojected to another coordinate system before using this
function. Note that for terra::rast() objects
boundaries are missing for cells that have missing (NA) values in all
cells.
Notes
In earlier versions, this function had an extra str_tree parameter
that could be used to leverage STR query trees to speed up processing
for planning units in vector format.
Although this functionality improved performance, it was not
enabled by default because the underlying function
(i.e., rgeos:gUnarySTRtreeQuery()) was documented as experimental.
The boundary_matrix() function has since been updated so that it will
use STR query trees to speed up processing for planning units in vector
format (using terra::sharedPaths()).
Also, note that in previous versions, cell values along the matrix diagonal indicated the perimeter associated with planning units that did not contain any neighbors. This has now changed such that values along the diagonal now correspond to the total perimeter associated with each planning unit.
See also
Boundary matrix data might need rescaling to improve optimization
performance, see rescale_matrix() to perform these calculations.
Examples
# \dontrun{
# load data
sim_pu_raster <- get_sim_pu_raster()
sim_pu_polygons <- get_sim_pu_polygons()
# subset data to reduce processing time
r <- terra::crop(sim_pu_raster, c(0, 0.3, 0, 0.3))
ply <- sim_pu_polygons[c(1:3, 11:13, 20:22), ]
# create boundary matrix using raster data
bm_raster <- boundary_matrix(r)
# create boundary matrix using polygon data
bm_ply <- boundary_matrix(ply)
# plot raster data
plot(r, main = "raster", axes = FALSE)
 # plot boundary matrix
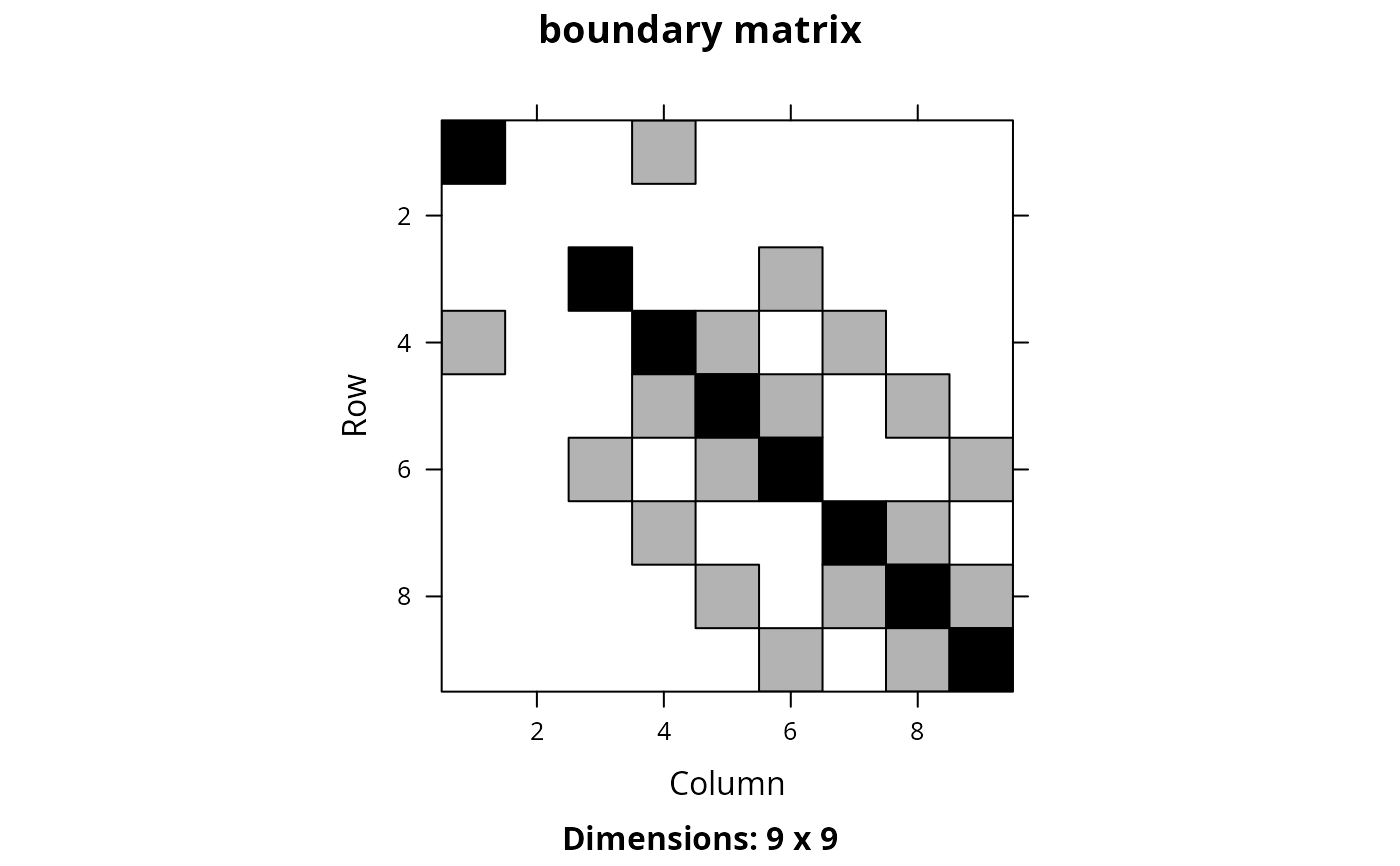
# here each row and column corresponds to a different planning unit
Matrix::image(bm_raster, main = "boundary matrix")
# plot boundary matrix
# here each row and column corresponds to a different planning unit
Matrix::image(bm_raster, main = "boundary matrix")
 # plot polygon data
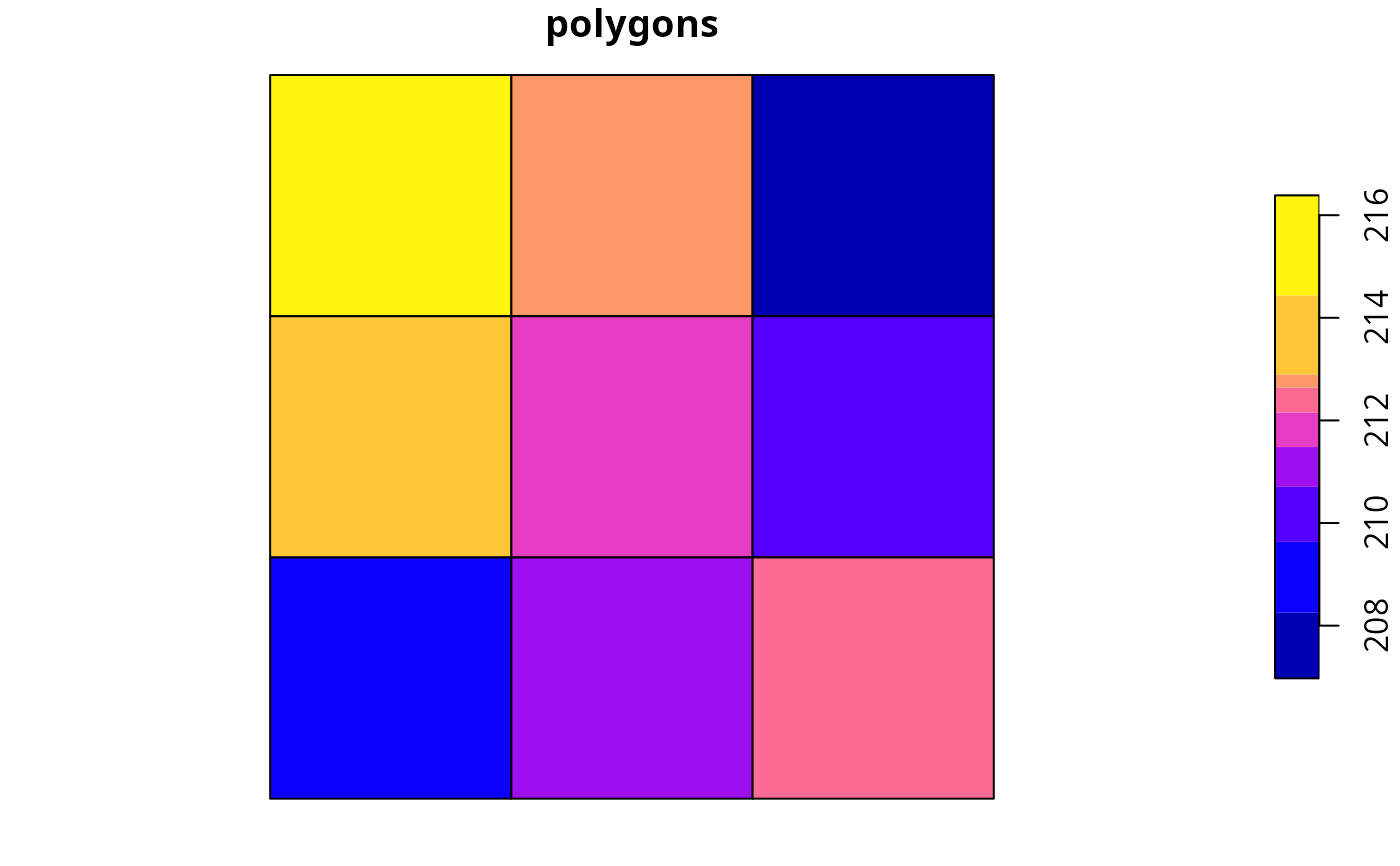
plot(ply[, 1], main = "polygons", axes = FALSE)
# plot polygon data
plot(ply[, 1], main = "polygons", axes = FALSE)
 # plot boundary matrix
# here each row and column corresponds to a different planning unit
Matrix::image(bm_ply, main = "boundary matrix")
# plot boundary matrix
# here each row and column corresponds to a different planning unit
Matrix::image(bm_ply, main = "boundary matrix")
 # }
# }
