
Specify targets following Watson et al. (2010)
Source:R/spec_watson_targets.R
spec_watson_targets.RdSpecify targets based on the methodology outlined by
Watson et al. (2010).
Briefly, this method involves setting targets thresholds as a percentage
based on whether or not a feature is considered rare.
To help prevent widespread features from obscuring priorities,
targets are capped following Butchart et al. (2015).
This method was designed for species protection at national-scales.
Note that this function is designed to be used with add_auto_targets()
and add_group_targets().
Usage
spec_watson_targets(
rare_area_threshold = 10000,
rare_relative_target = 1,
rare_area_target = 1000,
common_relative_target = 0.1,
cap_area_target = 1e+06,
area_units = "km^2"
)Arguments
- rare_area_threshold
numericvalue indicating the threshold area for rare identifying rare features. Defaults to 10000 (i.e., 10000 km^2).- rare_relative_target
numericvalue denoting the relative target for features with a spatial distribution that is smaller thanrare_area_threshold. Note that this value must be a proportion between 0 and 1. Defaults to 1 (i.e., 100%).- rare_area_target
numericvalue indicating the area-based target for features with a spatial distribution that is smaller thanrare_area_threshold. Defaults to 1000 (i.e., 1000 km^2).- common_relative_target
numericvalue denoting the relative target for features with a spatial distribution that is greater thancommon_area_threshold. Defaults to 0.1 (i.e., 10%). Since this default value is based on historical levels of global protected area coverage, it may be appropriate to set this value based on current levels of protected area coverage (e.g., 17.6% for terrestrial and 8.4% for marine systems globally; UNEP-WCMC and IUCN 2025).- cap_area_target
numericvalue denoting the area-based target cap. To avoid setting a target cap, a missing (NA) value can be specified. Defaults to 1000000 (i.e., 1,000,000 km2).- area_units
charactervalue denoting the unit of measurement for the area-based arguments. Defaults to"km^2"(i.e., km2).
Value
An object (TargetMethod) for specifying targets that
can be used with add_auto_targets() and add_group_targets()
to add targets to a problem().
Details
This target setting method was designed to protect species in
a national-scale prioritization (Watson et al. 2010).
Although similar methods have also been successfully to national-scale
prioritizations (e.g., Kark et al. 2009),
it may fail to identify meaningful priorities for
prioritizations conducted at smaller geographic scales
(e.g., local or county-level scales).
For example, if this target setting method is applied to
such geographic scales, then the resulting prioritizations
may select an overly large percentage of the study area,
or be biased towards over-representing common and widespread species.
This is because the thresholds
(i.e., rare_area_threshold and cap_area_threshold)
were originally developed based on criteria for national-scales.
As such, if you working at the sub-national scale, it is recommended to set
thresholds based on that criteria are appropriate to the spatial extent
of the planning region.
Please note that this function is provided as convenient method to
set targets for problems with a single management zone, and cannot
be used for those with multiple management zones.
Mathematical formulation
This method involves setting target thresholds based on the spatial
extent of the features.
By default, this method identifies rare features as those with a
spatial distribution smaller than 10,000
km2
(per rare_area_threshold and area_units)
and common features as those with a larger spatial distribution.
Given this, rare features are assigned target threshold of 100%
(per rare_relative_target)
or 1,000 km2
(per rare_area_threshold)
(whichever of these two values is smaller), and
common features are assigned a target threshold of 10%
(per common_relative_target).
Additionally, following Butchart et al. (2015), a cap of 1,000,000
km2 is applied to target
thresholds (per cap_area_threshold and area_units).
Data calculations
This function involves calculating targets based on the spatial extent
of the features in x.
Although it can be readily applied to problem() objects that
have the feature data provided as a terra::rast() object,
you will need to specify the spatial units for the features
when initializing the problem() objects if the feature data
are provided in a different format. In particular, if the feature
data are provided as a data.frame or character vector,
then you will need to specify an argument to feature_units when
using the problem() function.
See the Examples section of the documentation for add_auto_targets()
for a demonstration of specifying the spatial units for features.
References
Butchart SHM, Clarke M, Smith RJ, Sykes RE, Scharlemann JPW, Harfoot M, Buchanan GM, Angulo A, Balmford A, Bertzky B, Brooks TM, Carpenter KE, Comeros‐Raynal MT, Cornell J, Ficetola GF, Fishpool LDC, Fuller RA, Geldmann J, Harwell H, Hilton‐Taylor C, Hoffmann M, Joolia A, Joppa L, Kingston N, May I, Milam A, Polidoro B, Ralph G, Richman N, Rondinini C, Segan DB, Skolnik B, Spalding MD, Stuart SN, Symes A, Taylor J, Visconti P, Watson JEM, Wood L, Burgess ND (2015) Shortfalls and solutions for meeting national and global conservation area targets. Conservation Letters, 8: 329–337.
Kark S, Levin N, Grantham HS, Possingham HP (2009) Between-country collaboration and consideration of costs increase conservation planning efficiency in the Mediterranean Basin. Proceedings of the National Academy of Sciences, 106: 15368–15373.
UNEP-WCMC and IUCN (2025) Protected Planet Report 2024. Cambridge, UK: UNEP-WCMC and IUCN. Available at <www.protectedplanet.net>.
Watson JEM, Evans MC, Carwardine J, Fuller RA, Joseph LN, Segan DB, Taylor MFJ, Fensham RJ, Possingham HP (2010) The capacity of Australia's protected-area system to represent threatened species. Conservation Biology,25: 324–332.
See also
Other target setting methods:
spec_absolute_targets(),
spec_area_targets(),
spec_duran_targets(),
spec_interp_absolute_targets(),
spec_interp_area_targets(),
spec_jung_targets(),
spec_max_targets(),
spec_min_targets(),
spec_polak_targets(),
spec_pop_size_targets(),
spec_relative_targets(),
spec_rl_ecosystem_targets(),
spec_rl_species_targets(),
spec_rodrigues_targets(),
spec_rule_targets(),
spec_ward_targets(),
spec_wilson_targets()
Examples
# \dontrun{
# set seed for reproducibility
set.seed(500)
# load data
sim_complex_pu_raster <- get_sim_complex_pu_raster()
sim_complex_features <- get_sim_complex_features()
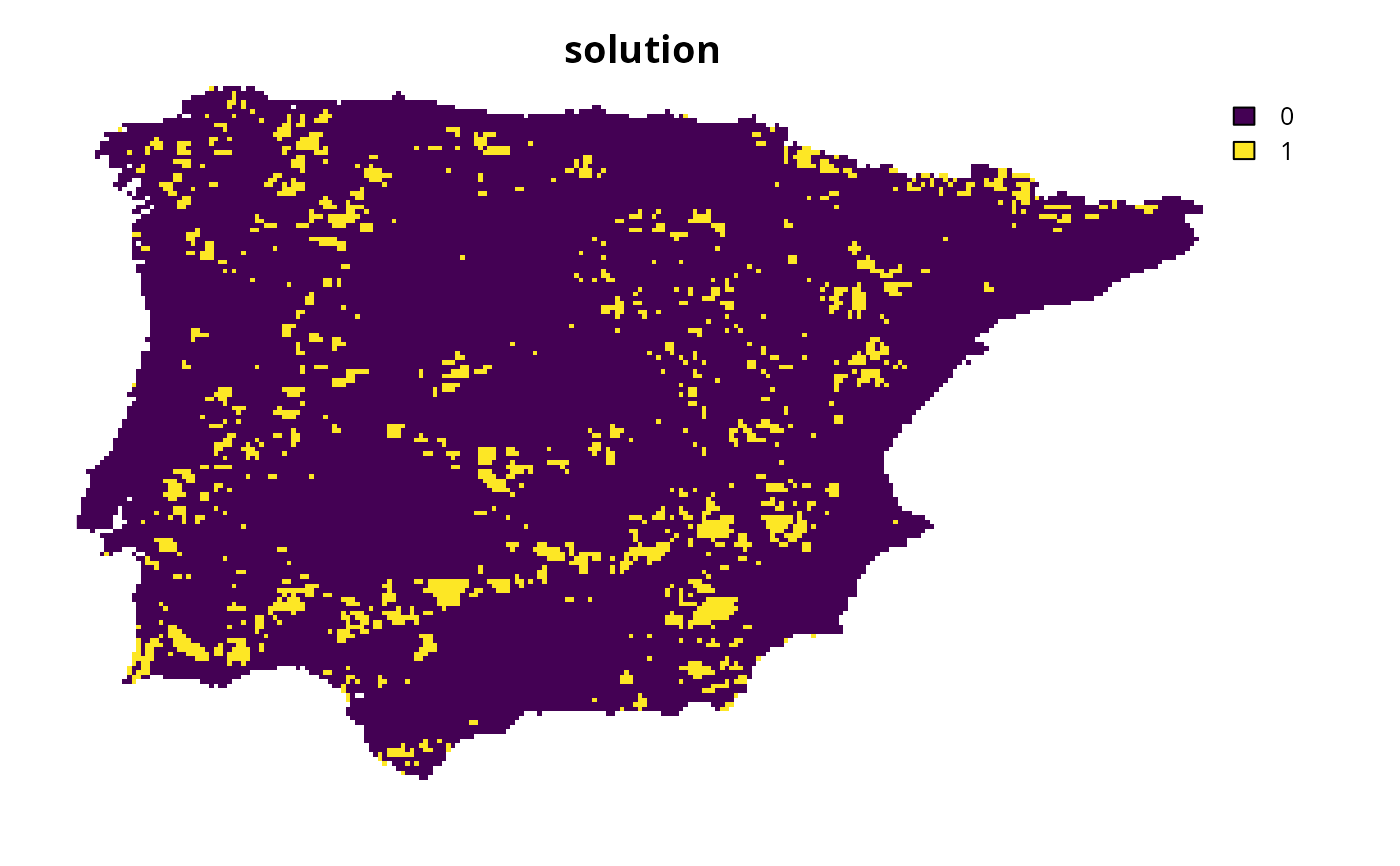
# create problem with Watson et al. (2010) targets
p1 <-
problem(sim_complex_pu_raster, sim_complex_features) %>%
add_min_set_objective() %>%
add_auto_targets(method = spec_watson_targets()) %>%
add_binary_decisions() %>%
add_default_solver(verbose = FALSE)
# solve problem
s1 <- solve(p1)
# plot solution
plot(s1, main = "solution", axes = FALSE)
 # }
# }